Achieving the goals of the MDLMA project requires substantial compute resources, and easy access to the resources, data and related services. Some services are tailored for non-experts to foster sustainability.
Services
- Maxwell: HPC for MDLMA and beyond
- JupyterHub: Custom kernels for MDLMA
- Share a Scope: Collaborative platform (*)
- SciCat frontend - Visualizing metadata
- SLURM configurator - Compose, submit and view batch-jobs
- MLexchange: Machine Learning pipeline for tomography (*)
- WebJobs: Live monitoring of job performance (*)
- MDLMA SciCat: Datamanagement platform
- Not a binder: Open Data Demonstrator (*)
(*) Currently reachable only inside the DESY network
Resources
The MDLMA has a dedicated partition (mdlma) in the Maxwell Cluster. The partition holds a single node with 4*V100 GPGPUs and 1.5TB of ram, funded by the project. The node despite being dedicated to the MDLMA project can be used by all Maxwell users in a parasitic manner (preemption) to maximize utilisation of the resource. MDLMA project members can in return use the entire cluster in the same way, to offer as many resources as needed. The MDLMA project has indeed consumed significantly more resources than offered by a single node benefiting from Maxwells collaborative concept.
Share a Scope
Accessing SciCat, running batch-jobs or view the results of a deep learning from simple python scripts requires not only a bit of coding but also knowledge about various APIs like for example the scicat API or the slurm REST API. Submitting batch jobs is simple enough, but to make a good choise of parameters for a particular ML/DL is less obvious without a good knowledge of SLURM and the Maxwell cluster.
We’ve implemented a variation of the CDS dashboard, which allows to provide very simple interfaces and templates to users. max-sas also enables sharing of arbitrary dashboards with others, which can save a lot of time to implement your own solution.
Each individual dashboard is a seperate web-service running as a batch job on a dedicated slurm cluster. The dashboards are running entirely in user space, which provides full access to all data in a controlled manner.
 To demonstrate the capabilities we’ve implemented a small number of template dashboards:
To demonstrate the capabilities we’ve implemented a small number of template dashboards:
SciCat frontend
SciCat is designed to hold metadata for beamtimes and datasets. The raw or processed data are however inaccessible from SciCat, except for snapshots uploaded as attachments to metadata. The support for filetypes is essentially limited to images. Support for HDF5/Nexus files, videos and similar media files has to be implemented in SciCat frontend, which is a major effort.
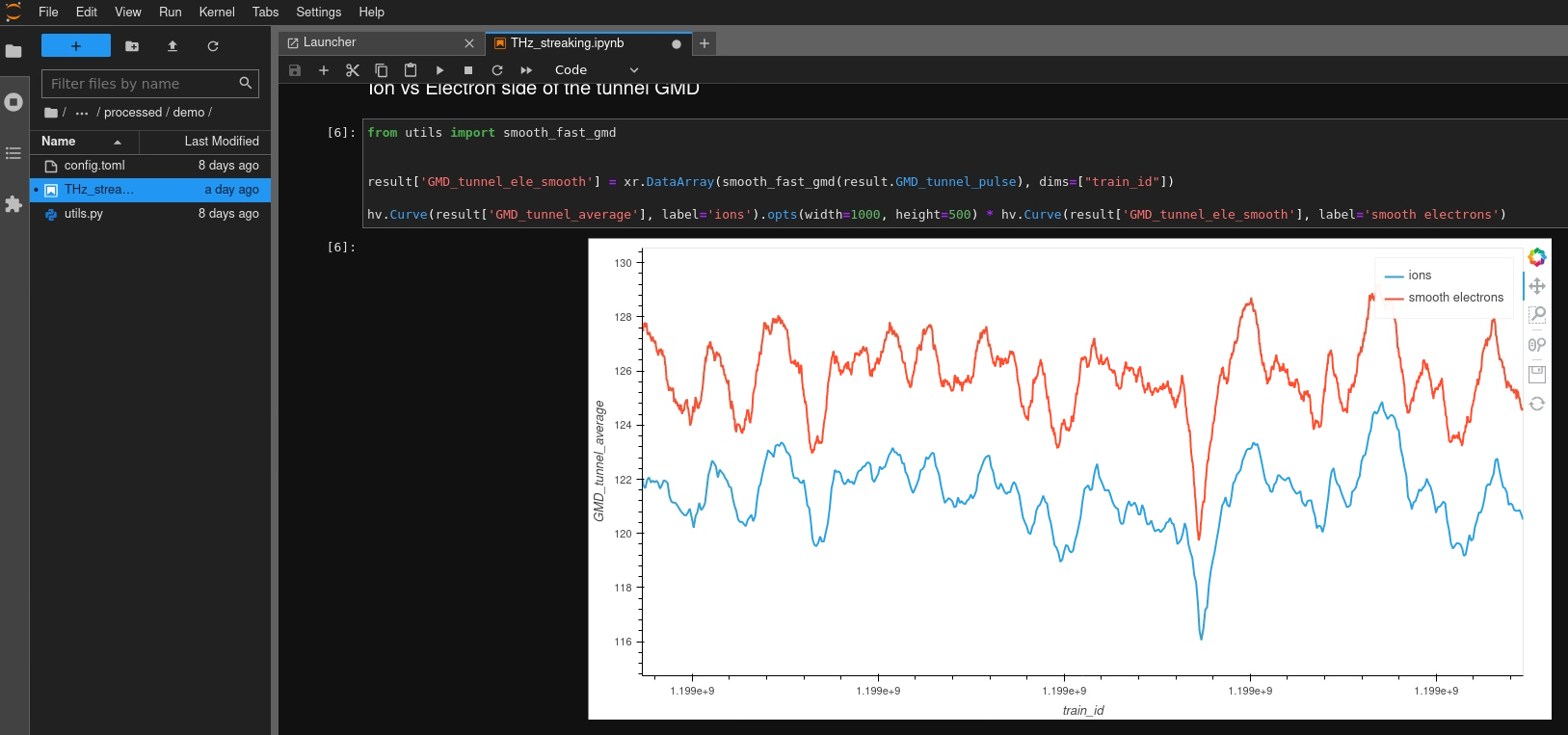
We therefore implemented a dashboard with reach features which also includes supports various file formats with just a few lines of python code:
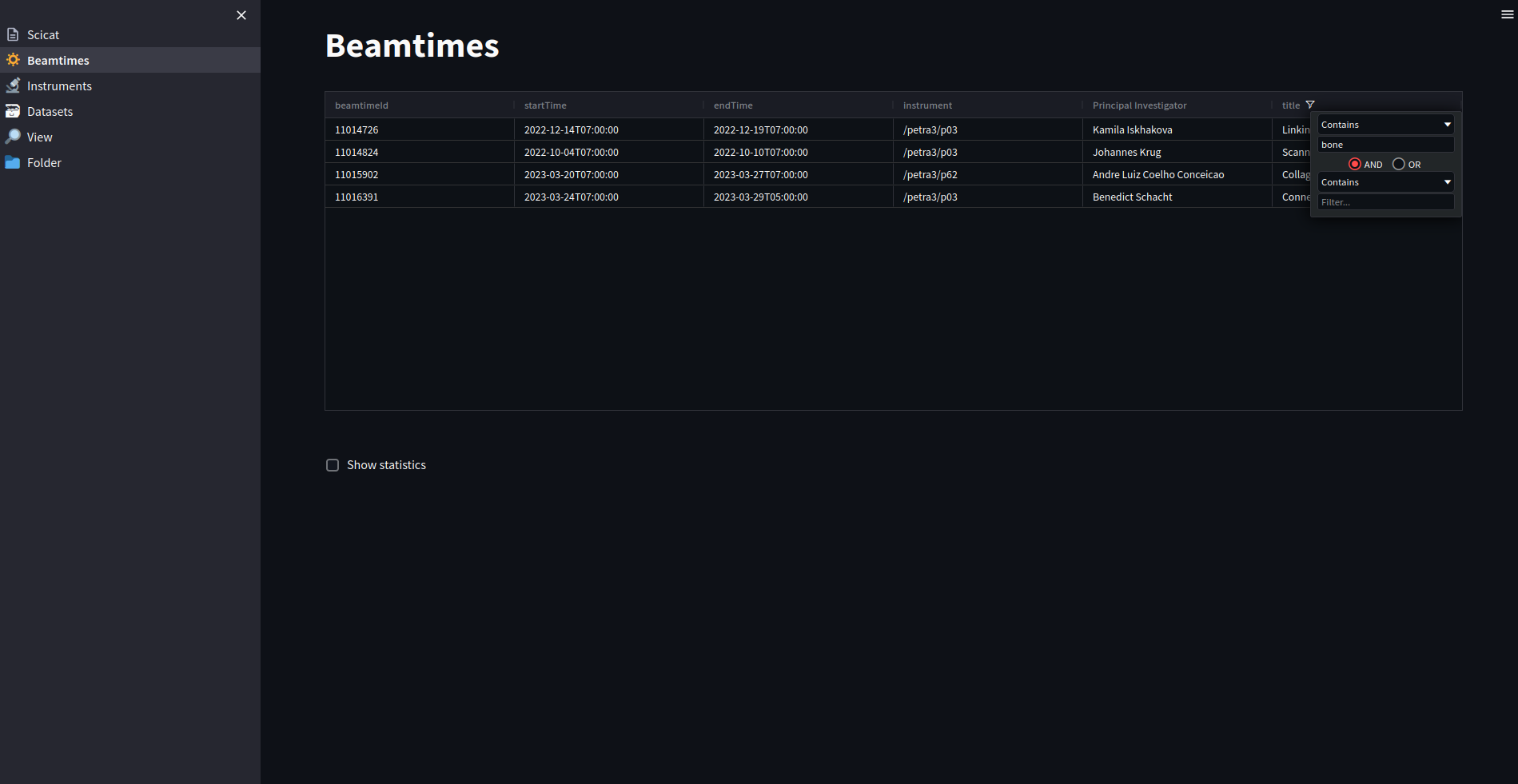
The dashboard exhibits all beamtimes and dataset a user is allowed to access. It does not implement a full search across all scientific metadata - that’s what SciCat is good for - but focusses on a few items like the title, which allows for instanteneous display of search results.
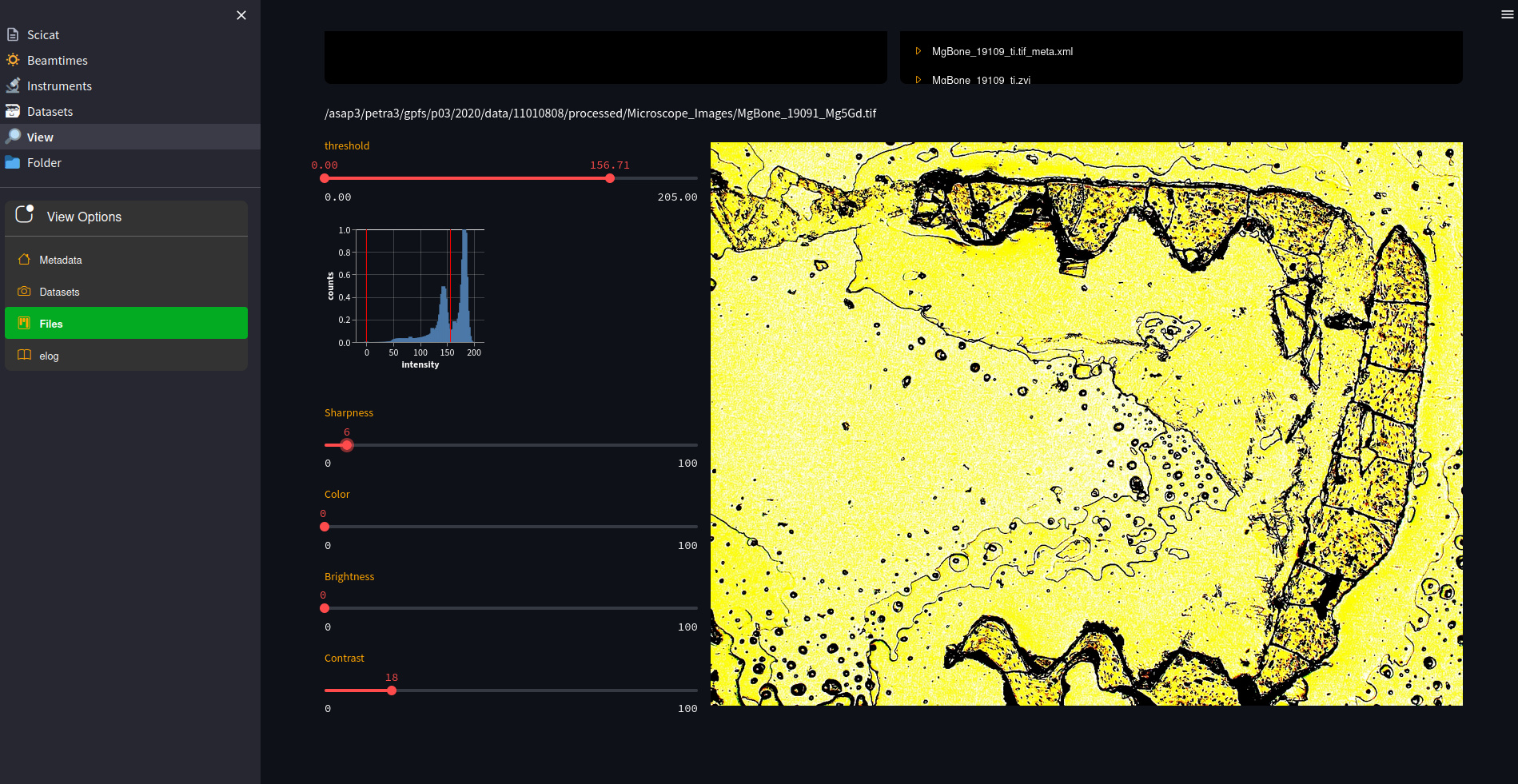
Since the dashboard is running in the users context, it has access to all files. A small fileselector allows to traverse the entire beamtime folder and visualize contents on the fly and in a customizable way.
All information is obtained from SciCat through its REST API with the access token obtained at login to the dashboard system. The user has hence direct access without any additional operation or knowledge.
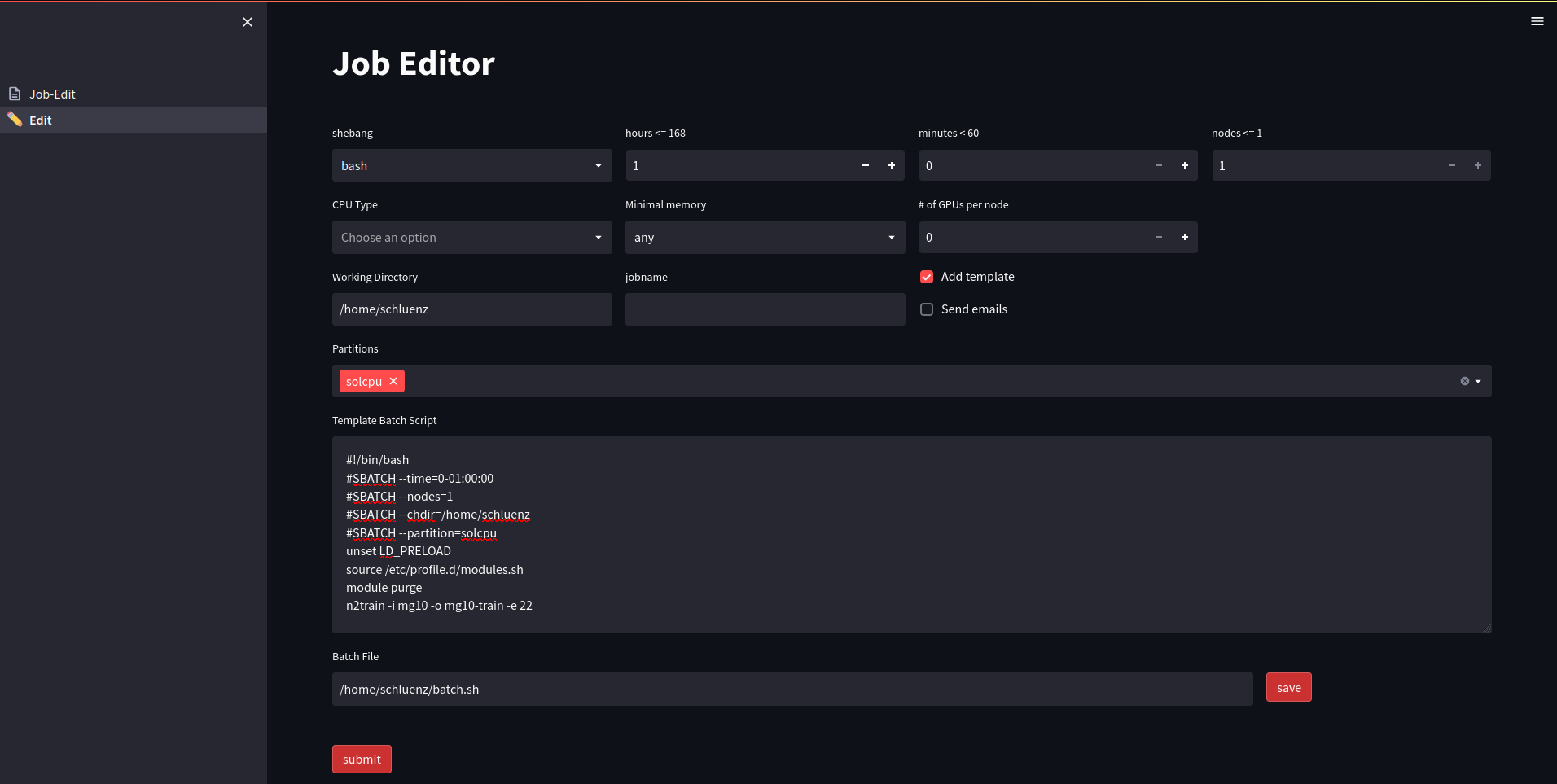
SLURM configurator
Designing a slurm batch script is generally simple enough, but users are frequently struggling selecting the proper set of partitions and constraints for a particular task. We therefore designed a dashboards which automatically detects allowed partitions, possible sets of constraints and timelimits. It composes a job-script which can be submitted directly with a single click, while the corresponding SLURM REST API token was automatically generated at login. The dashboard can easily be templated to set all variables for example for tasks like denoising or segmenting images. One could go a step further and connect it directly to SciCat, so that a user can select a beamtime and the tasks to be performed, but that has not been implemented yet.
Data visualization
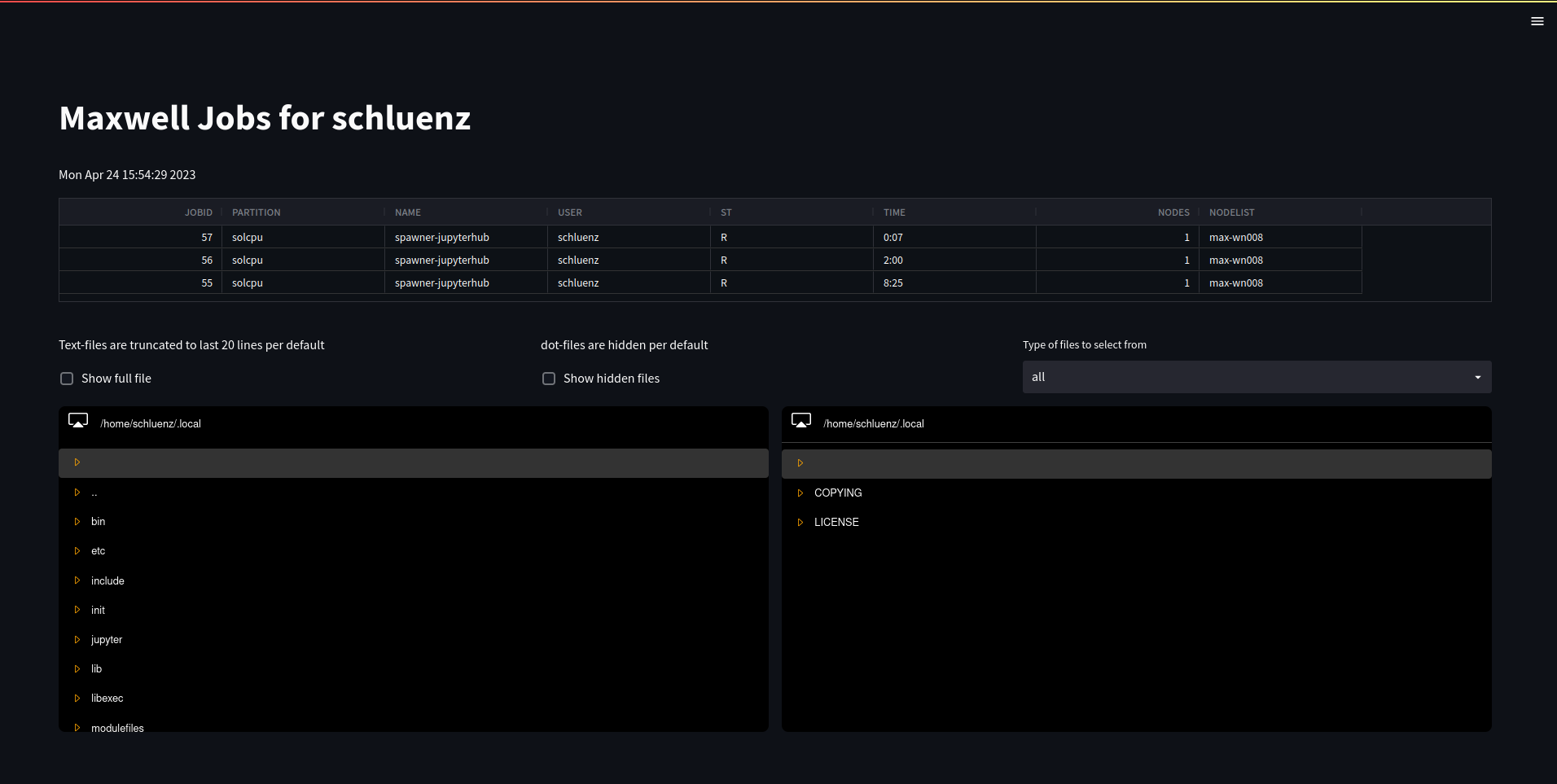
The Maxwell cluster offers full graphical access via fastx client or webbrowser for all users including of course members of the MDLMA project. It’s hence straightforward to monitor progression of e.g. machine learning tasks in real time. The graphical login is however naturally unaware of the batch system, so one needs keep track of batch jobs and related storage locations.
The dashboard system offers a direct view on (running) batch-jobs and is aware of locations and output files. It hence allows a very user friendly way to visualize jobs, logs and outputs of batch jobs.
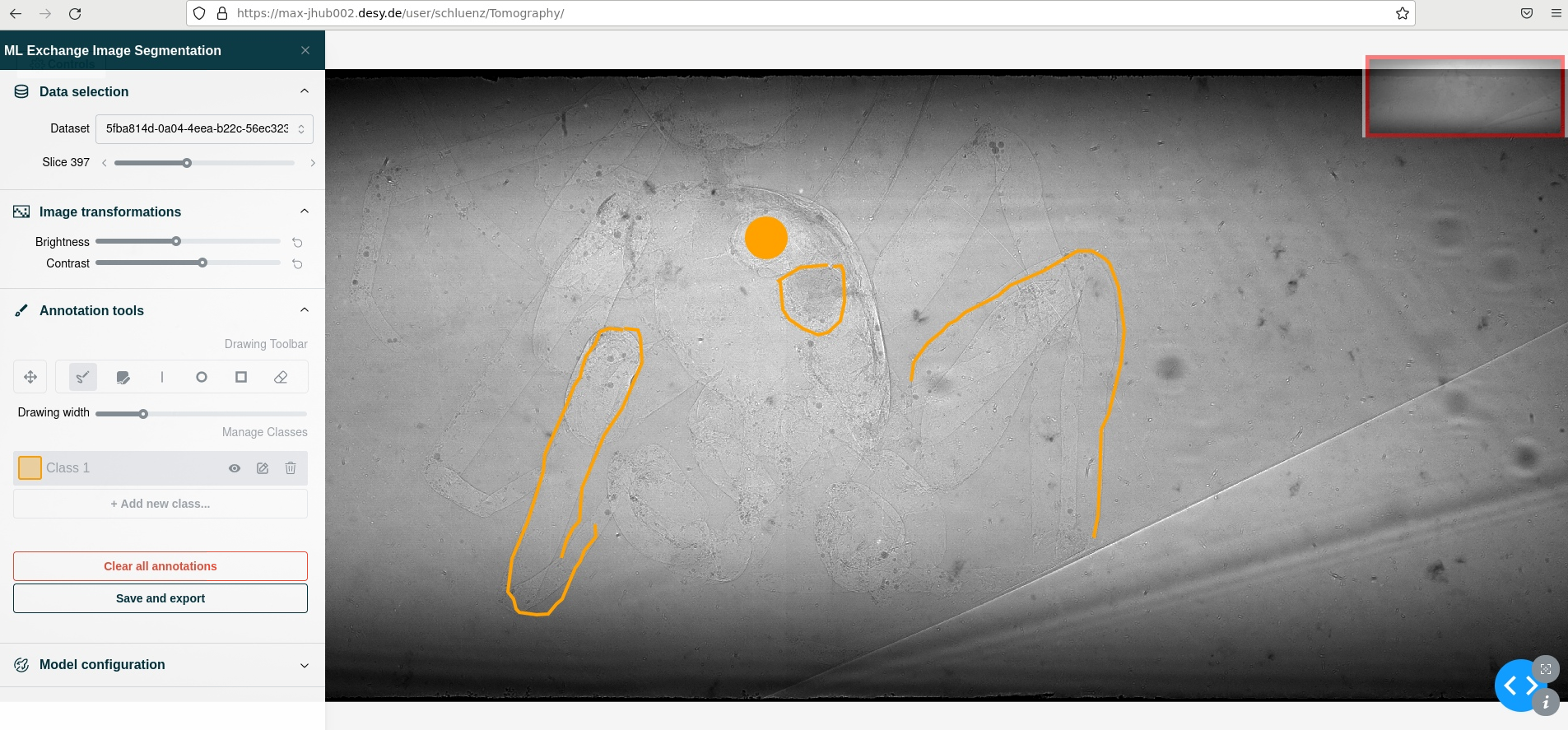
MLexchange
MLexchange implements a machine learning pipeline tailored for tomography applications, but is actually designed to be very generic. MLexchange is under development and uses BlueSky components still considered experimental. We nevertheless implemented part of the MLexchange pipeline as part of the SAS platform. The application is launched by and proxied through a customized JupyterHub instance. This SLURM batch job initiates a BlueSky TiledDB serving one or several datasets to the MLexhange annotation service. All components are executed in a user context, which makes the application quite secure, thereby providing a very simple data access rights management. The platform can also serve quite well for citizen science approaches.
The snapshot shows a sketch of an animation for an amber embedded spider.
JupyterHub
The Maxwell jupyterHub is available to the MDLMA project, and allows very simple access to substantial compute resources. To facilitate the MDLMA computations a number of typical applications are available as standard jupyter kernels. The kernels refer to complete environments and work without any additional installations or customizations.

Not a binder
BinderHub is a popular platform to allow anonymous execution of almost arbitrary workflows. Binder uses repo2docker as its core component, which is however not exactly matching our and our users needs. We have hence implemented an alternative solution. A systemd service serves a streamlit based entry point running inside a container. The container allows to select one of the configured open datasets. The configuration defines the workflows to be executed.
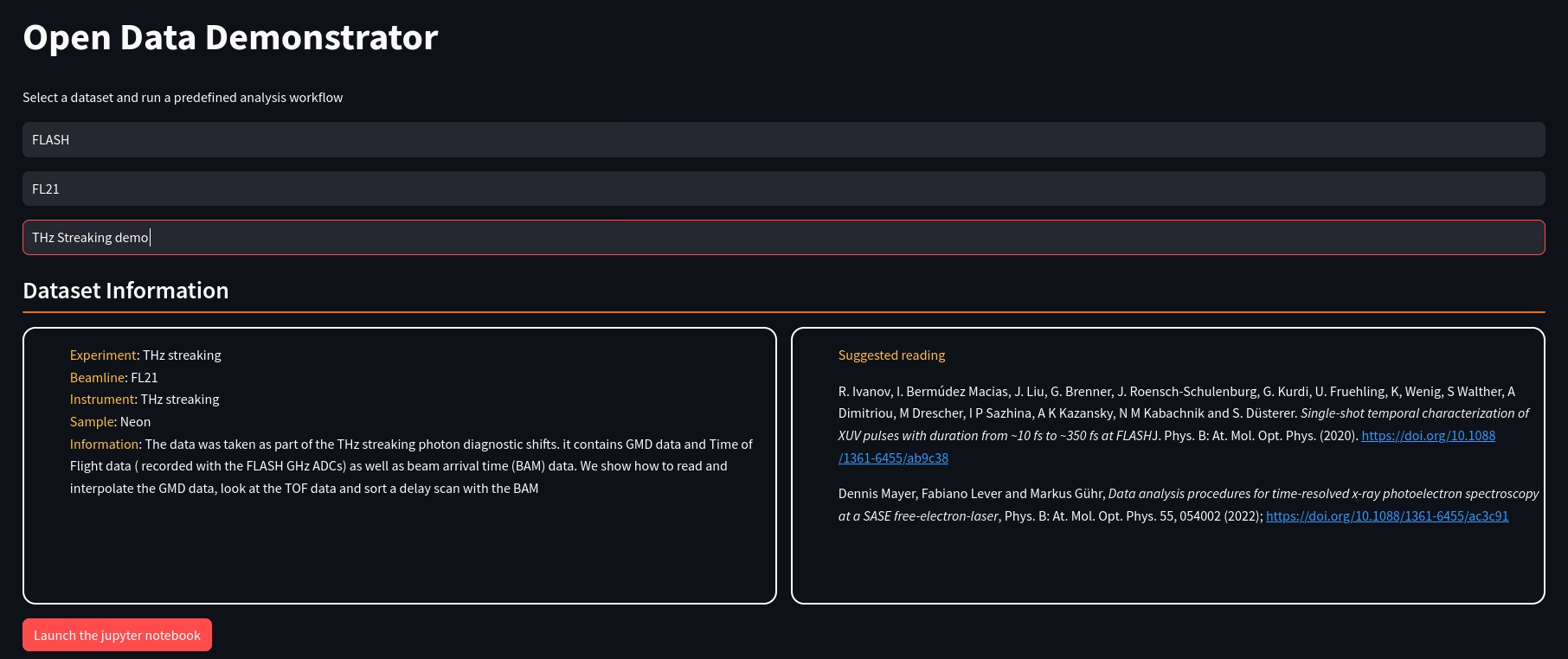
In this particular example the workflow is encoded in a jupyter notebook. The configuration defines the notebook, the kernel and their location in the filesystem. The submit button launches a SLURM batch job, which instantiates a container running a dedicated jupyter notebook server through the jupyterhub REST API while mounting all the ingredients necessary to execute the notebook. The container isolates the anonymous user from all non-essential resources, and the REST API is protected from external access, thereby providing a highly secure environment.